
Investigators
- Dr. Jessica Archibald - Radiology, Weill Cornell Medicine, New York, USA.
- Dr. Alexander Mark Weber - Pediatrics, University of British Columbia, Vancouver, Canada.
- Dr. Paulina S. Scheuren - Anesthesiology, Pharmacology, and Therapeutics, University of British Columbia, Vancouver, Canada.
- Oscar Ortiz - MSc, Research Technician, ICORD, Vancouver, Canada.
- Jaime Lee - BSc, Anesthesiology, Pharmacology, and Therapeutics, University of British Columbia, Vancouver, Canada
- Cassandra Choles - MSc, Anesthesiology, Pharmacology, and Therapeutics, University of British Columbia, Vancouver, Canada.
- Dr. Niklaus Zölch - Psychiatry and Forensic Medicine, Universität Zürich, Zürich, Switzerland.
- Dr. Erin L. MacMillan* - Radiology, University of British Columbia, Vancouver, Canada. ImageTech Lab, Simon Fraser University, Surrey, Canada. Philips Healthcare Canada, Markham, Canada.
- Dr. John L. K. Kramer* - Anesthesiology, Pharmacology, and Therapeutics, University of British Columbia, Vancouver, Canada.
Funding


How to cite this work
If you find this work helpful or utilize any part of it in your research or publications, please consider citing it as follows:
J. Archibald, et.al. Integrating Structural, Functional, and Biochemical Brain Imaging Data with MRShiny Brain - An Interactive Web Application. [Journal]. [Year of Publication]. [URL or DOI].
Welcome to the Study Description page!
This is where you can read more about our study and its design.
Participants: Healthy individuals were invited to participate in the study and were asked to come to the laboratory in a fasting state.
The timing of the scan was kept consistent (11:30am-12:30pm) across participants (to account for circadian rhythm effects of metabolites [Eckel-Mahan et al Physiol Rev 2013]).
Women were tested approximately during the follicular phase of their cycle [Hjelmervik, H Neuroimage 2018].
MR Procedures:
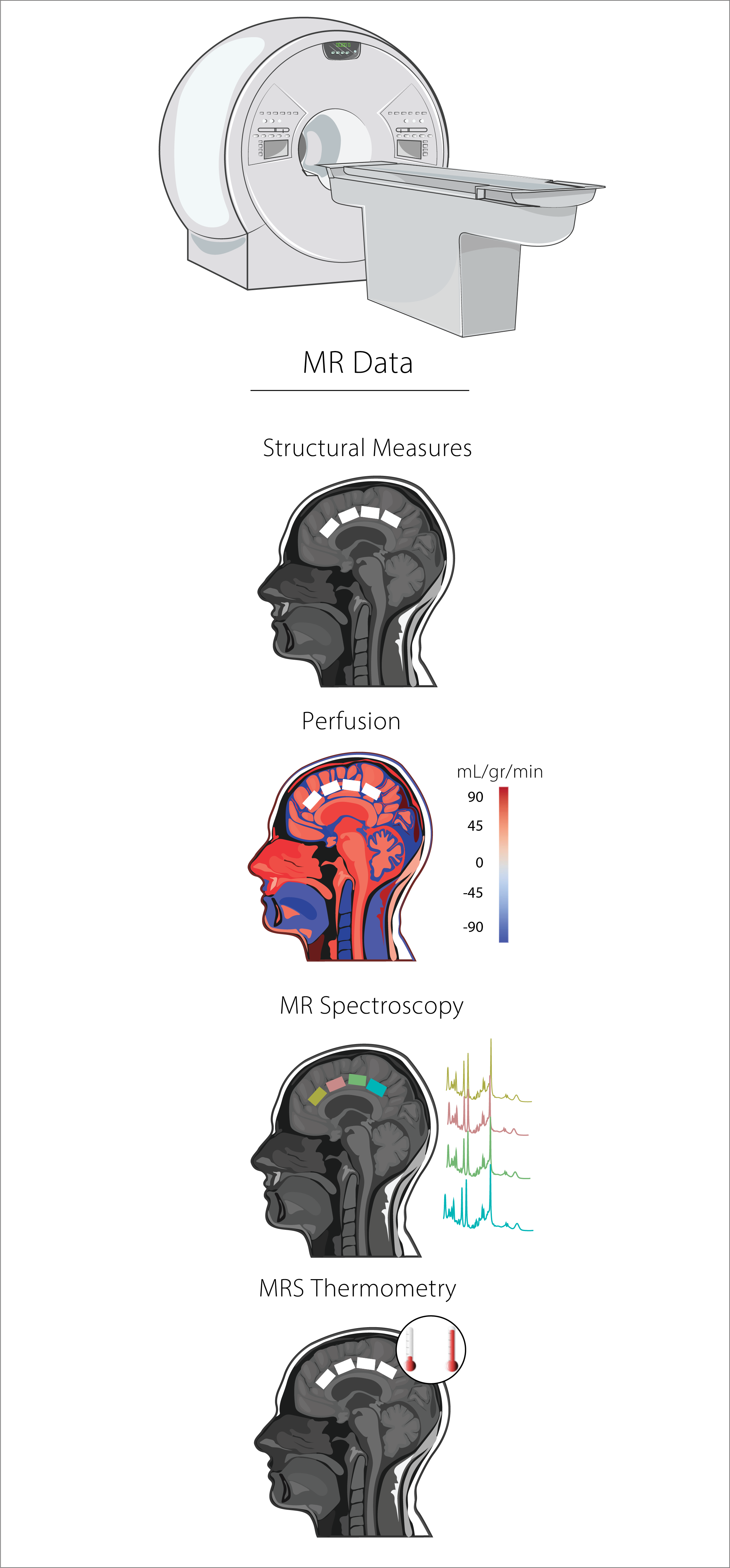
MRI data were collected using a 3T Philips Ingenia Elition X with a 32-channel SENSE head coil, and the sequences included:
- MPRAGE 3D T1 (TE/TR/TI=4.3/9.3/950ms, shot interval=2400ms, +0.8mm³ isotropic resolution, FOV (ap/rl/fh)=256/256/180mm³, scan time=5:49).
- A pseudo-continuous arterial spin labelling (pCASL) sequence was used to assess CBF. The sequence consisted of 4 pairs of perfusion-weighted and control scans (TE/TR=12/4174ms, post-labelling duration=2000ms, labelling duration=1800, total scan duration=5.59 min).
- Single voxel 1H-MRS (sLASER, TE/TR=32/5000ms, NSA=64, voxel size=24/22/15mm³ =7.9mL, automated 2nd order shimming, 32-step phase cycle, water suppression using the frequency selective Excitation option). The order of MRS acquisition in the voxels was randomized for each participant.
MR Analysis:
Structural Measures
Image Segmentation was performed in FSL (v 6.05) using default options, ROI segmentation was performed using in-house MATLAB scripts. ROI Cortical Thickness was performed in native space for each subject using Freesurfer (v 7.2.0) - Code availability.
Perfusion
ROI perfusion levels were extracted in native space using ASLprep and FSL - Code availability.
MR Spectroscopy
MRS data was pre-processed (e.g., frequency alignment, and eddy-current correction) and quantified using in-house MATLAB scripts. Spectral fitting was performed in LCModel (6.3). The basis set was simulated using FID-A functions. Simulated metabolites included: Ala, Asc, Asp, Cre, GABA, Gln, Glc, Glu, Gly, GPC, GSH, Ins, Lac, NAA, NAAG, PCr, PE, Scyllo, and Tau - Code availability.
MRS Thermometry
After data pre-processing (i.e., frequency alignment, eddy current correction), local brain temperature (TB) was estimated by calculating the chemical shift difference between water and NAA measured in parts per million (ppm) using the following equation: